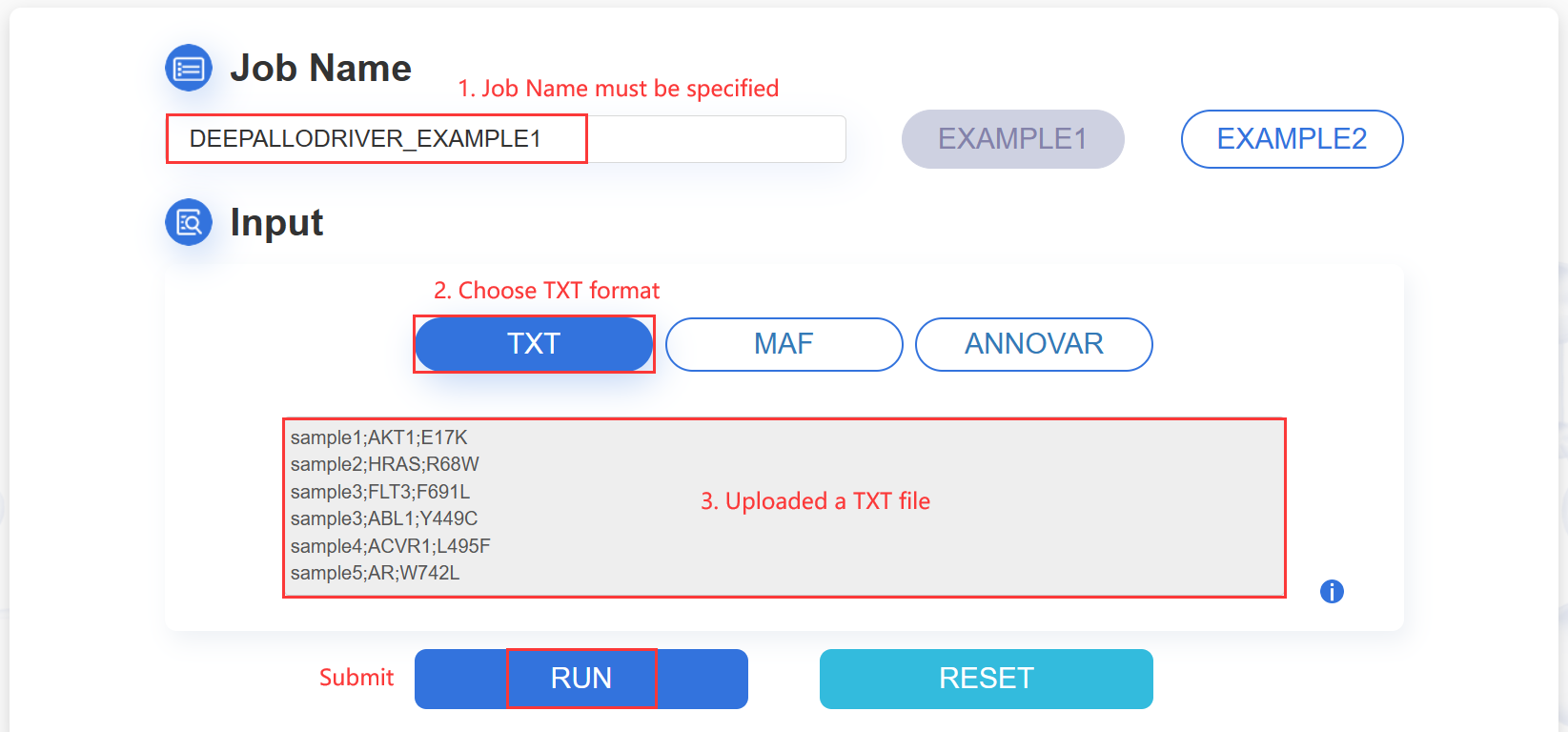
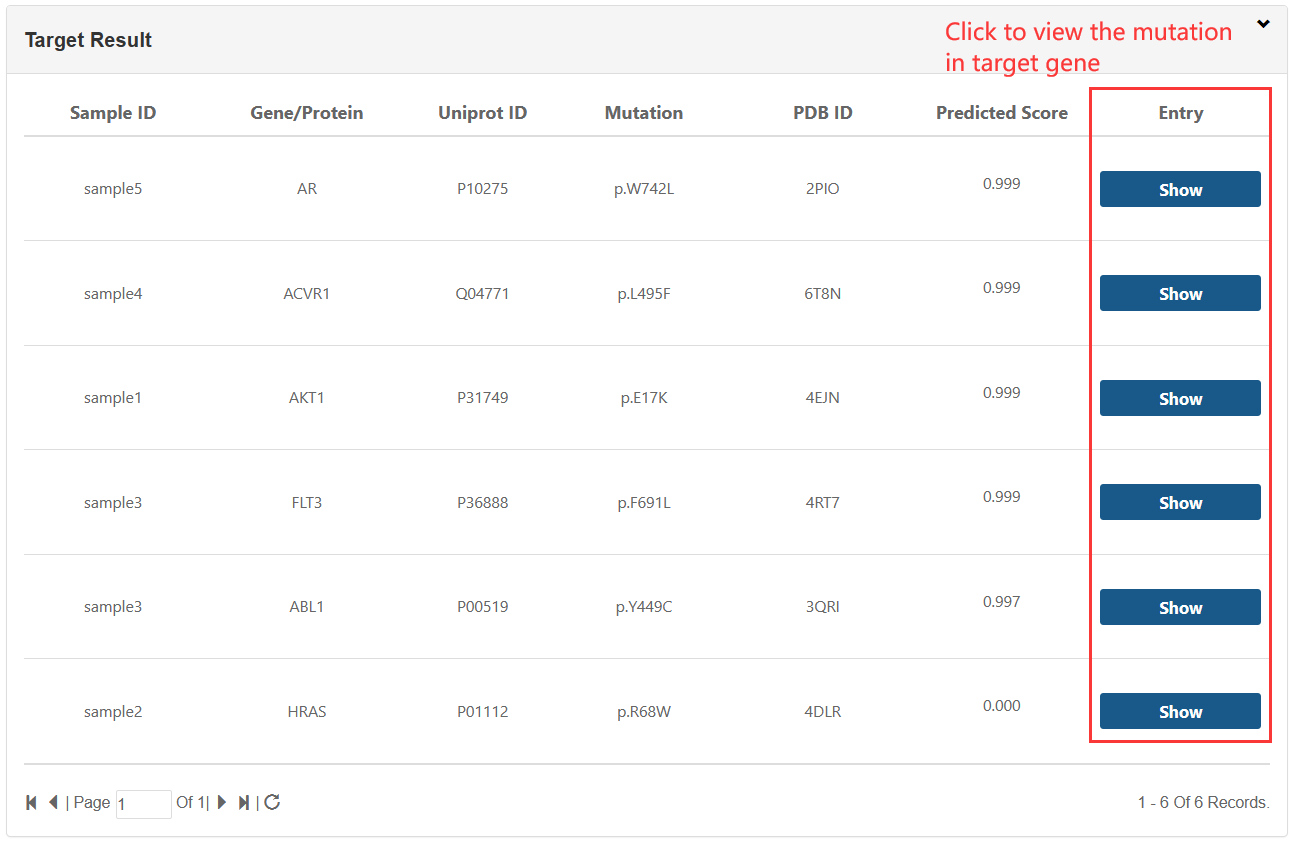
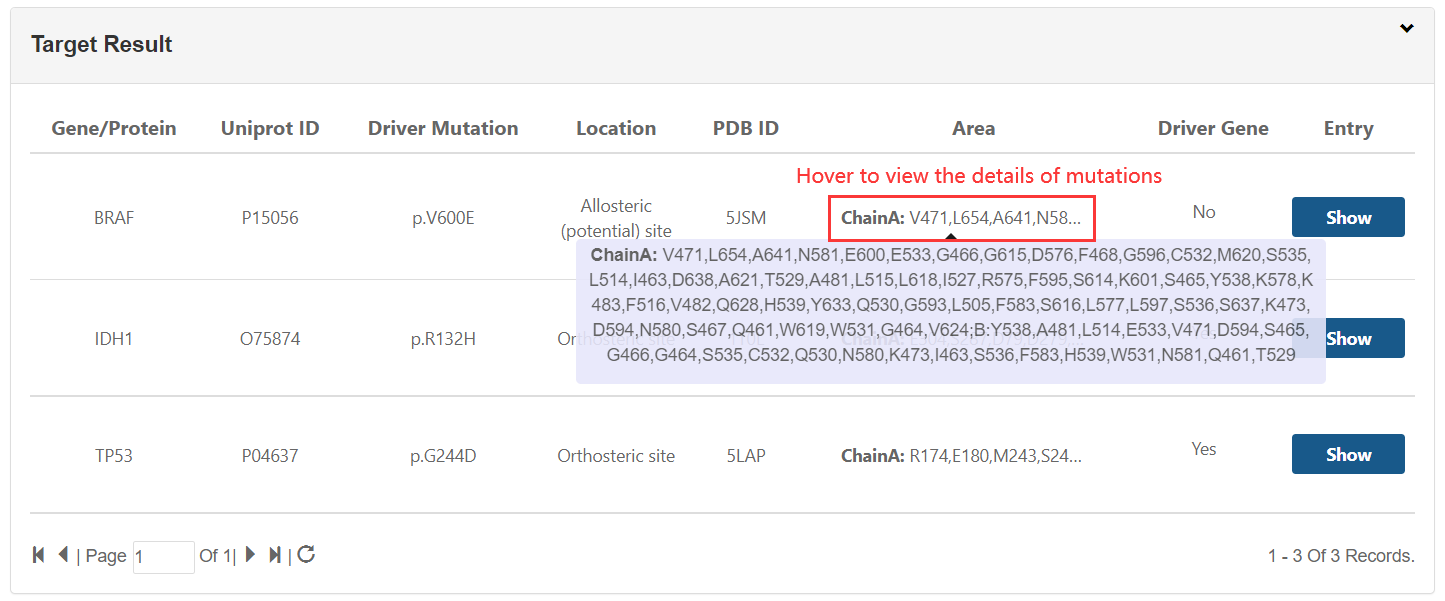
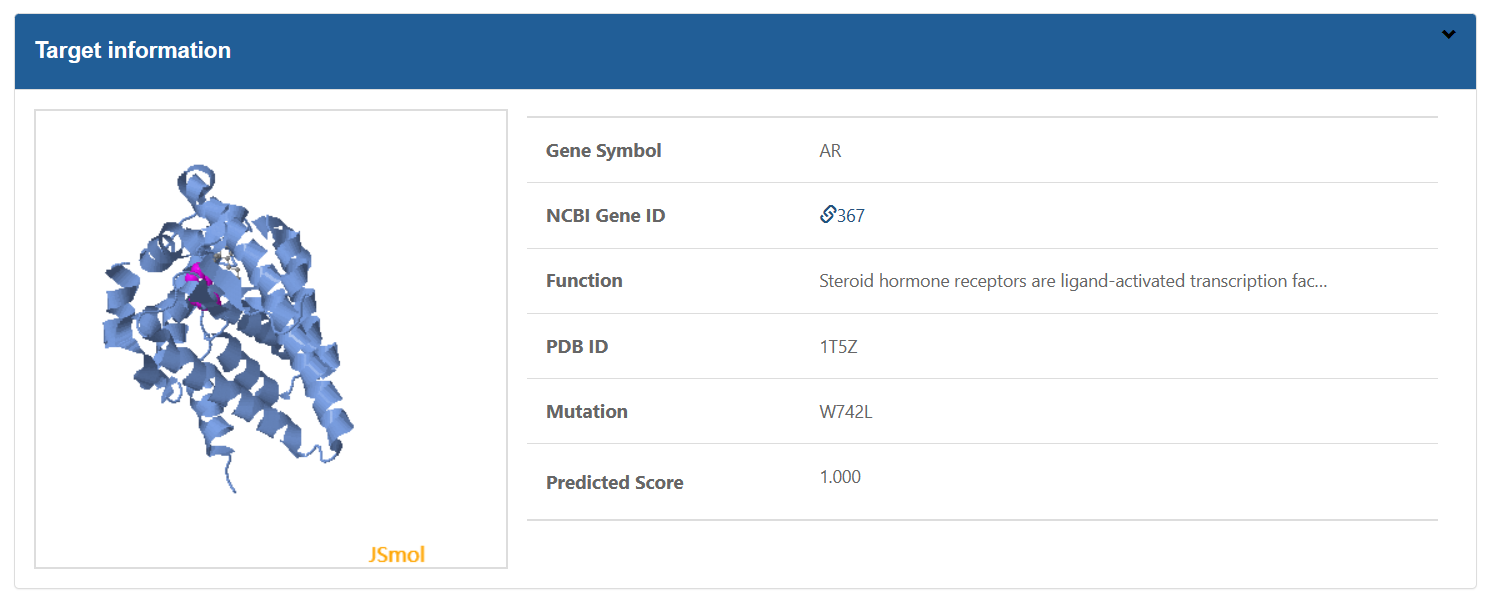
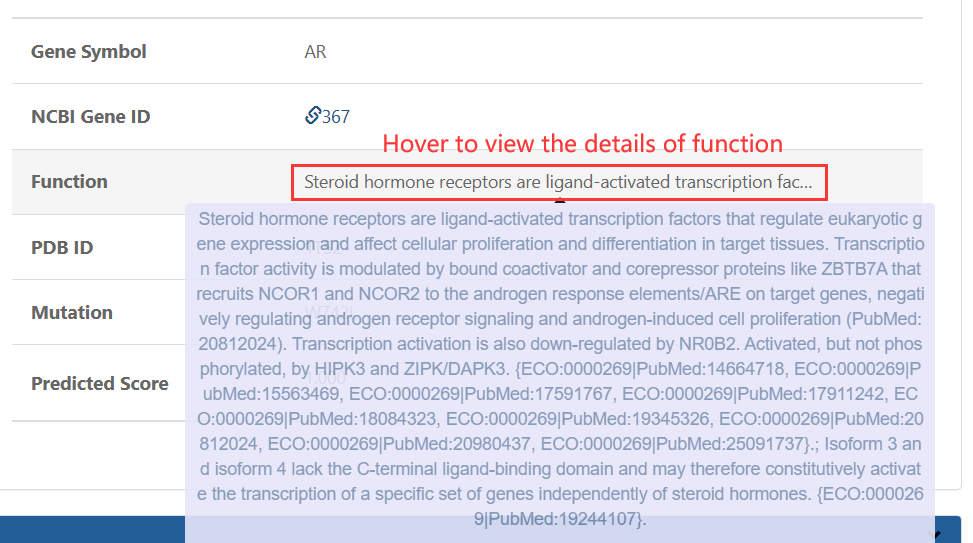
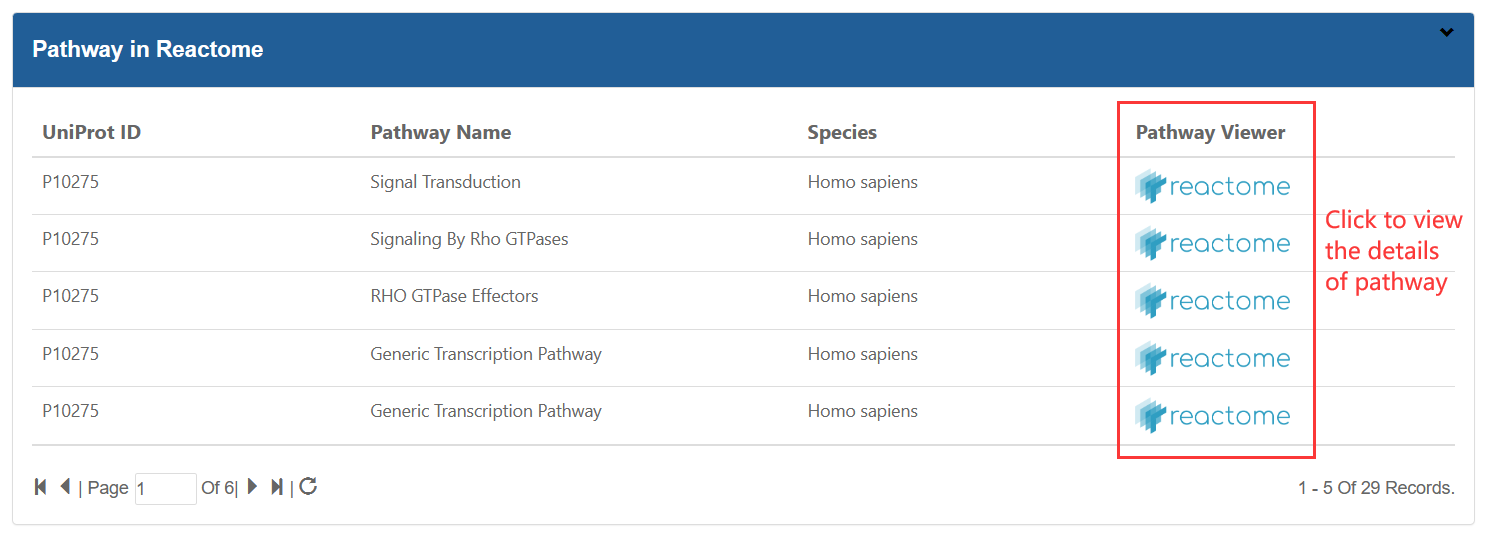
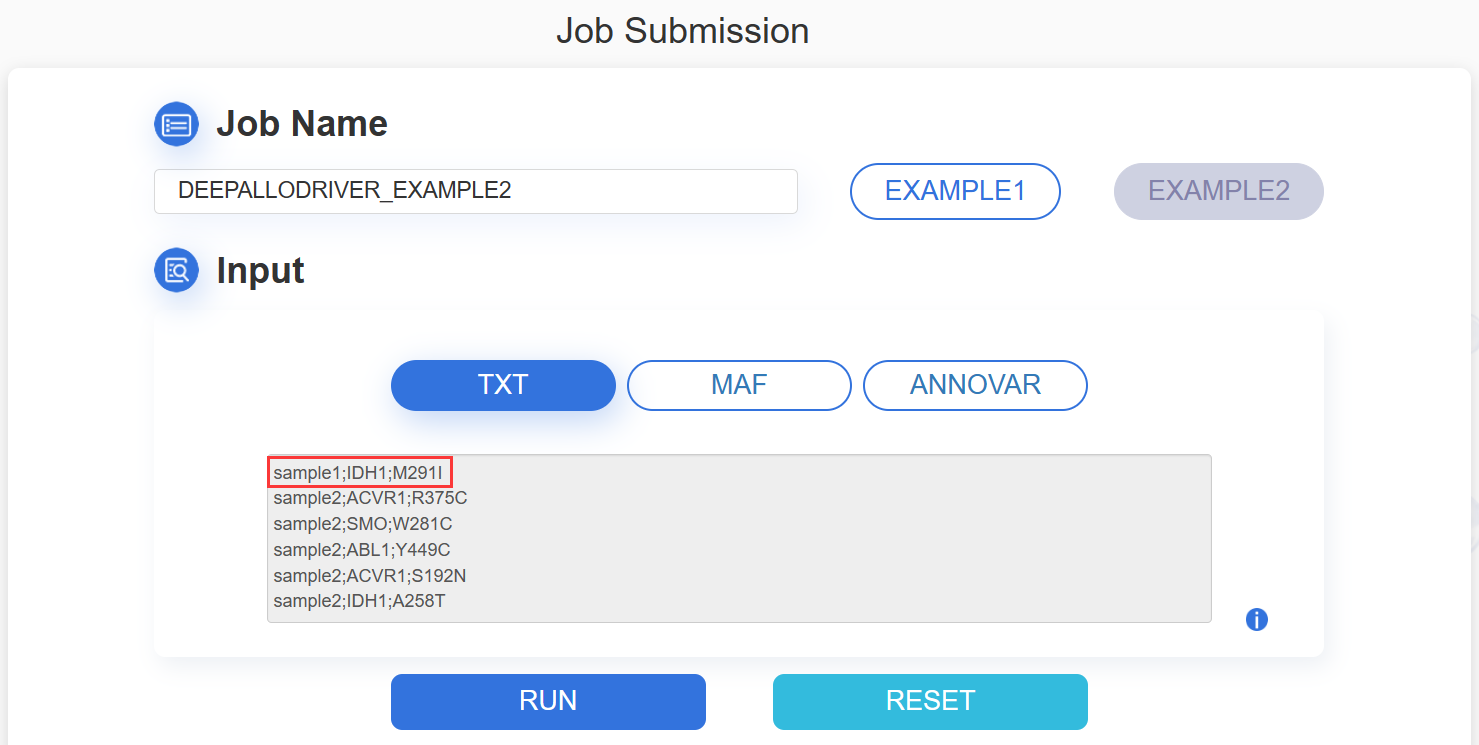
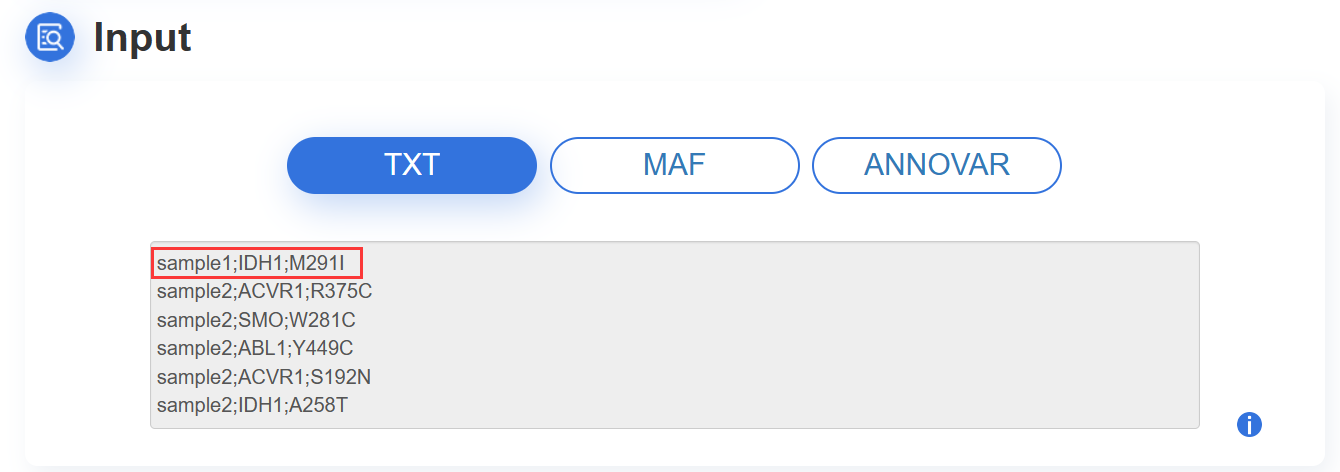
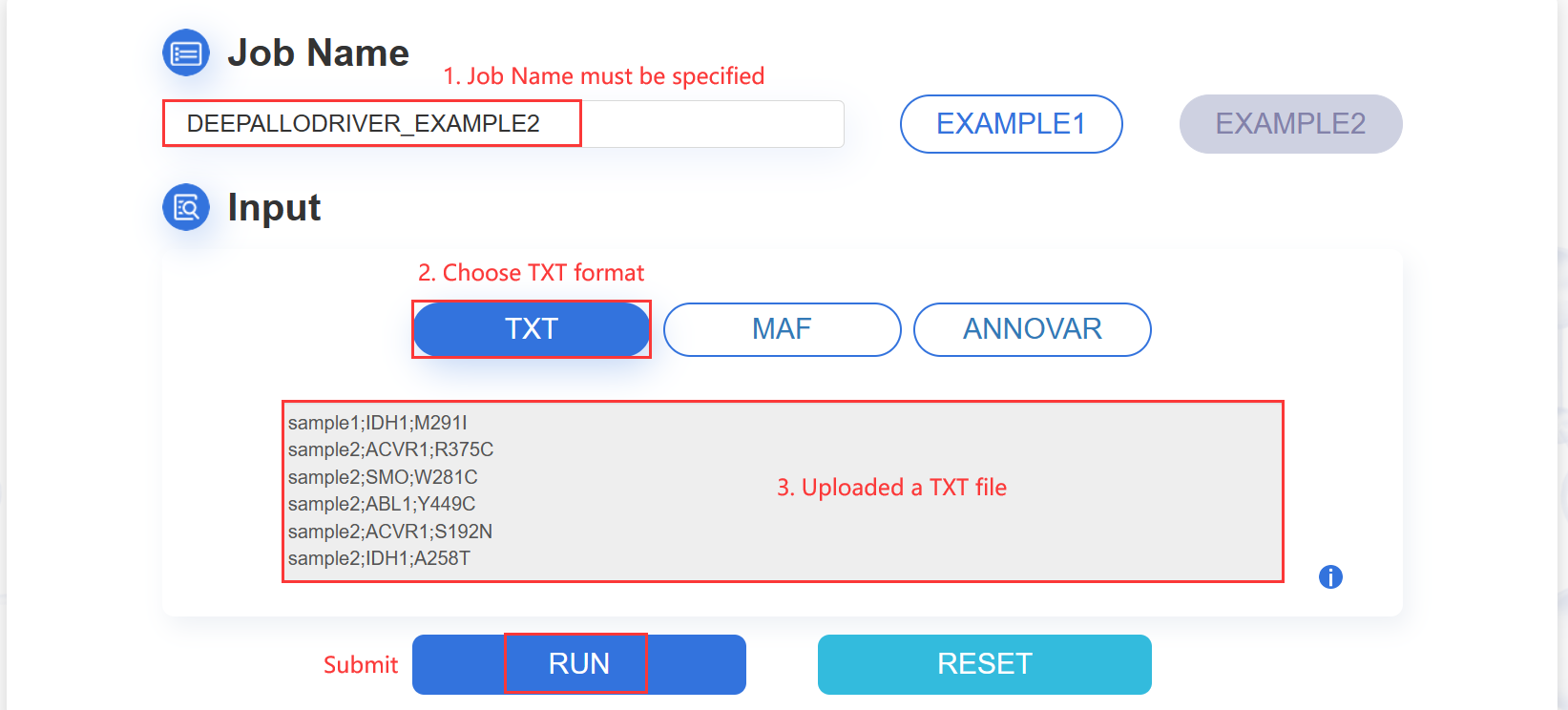
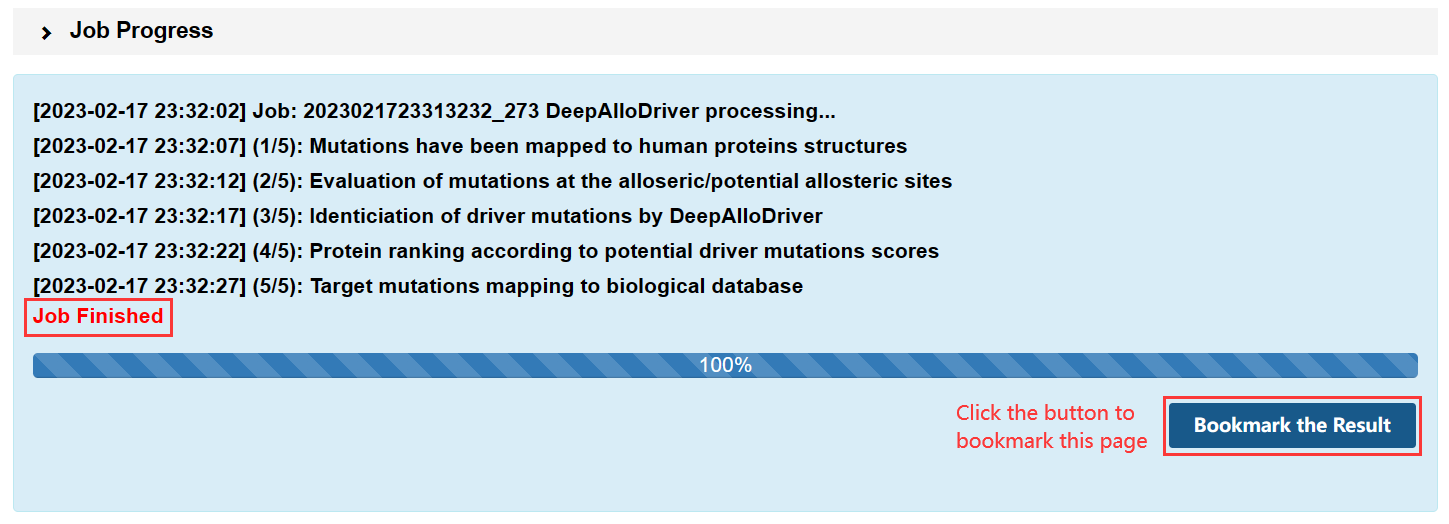
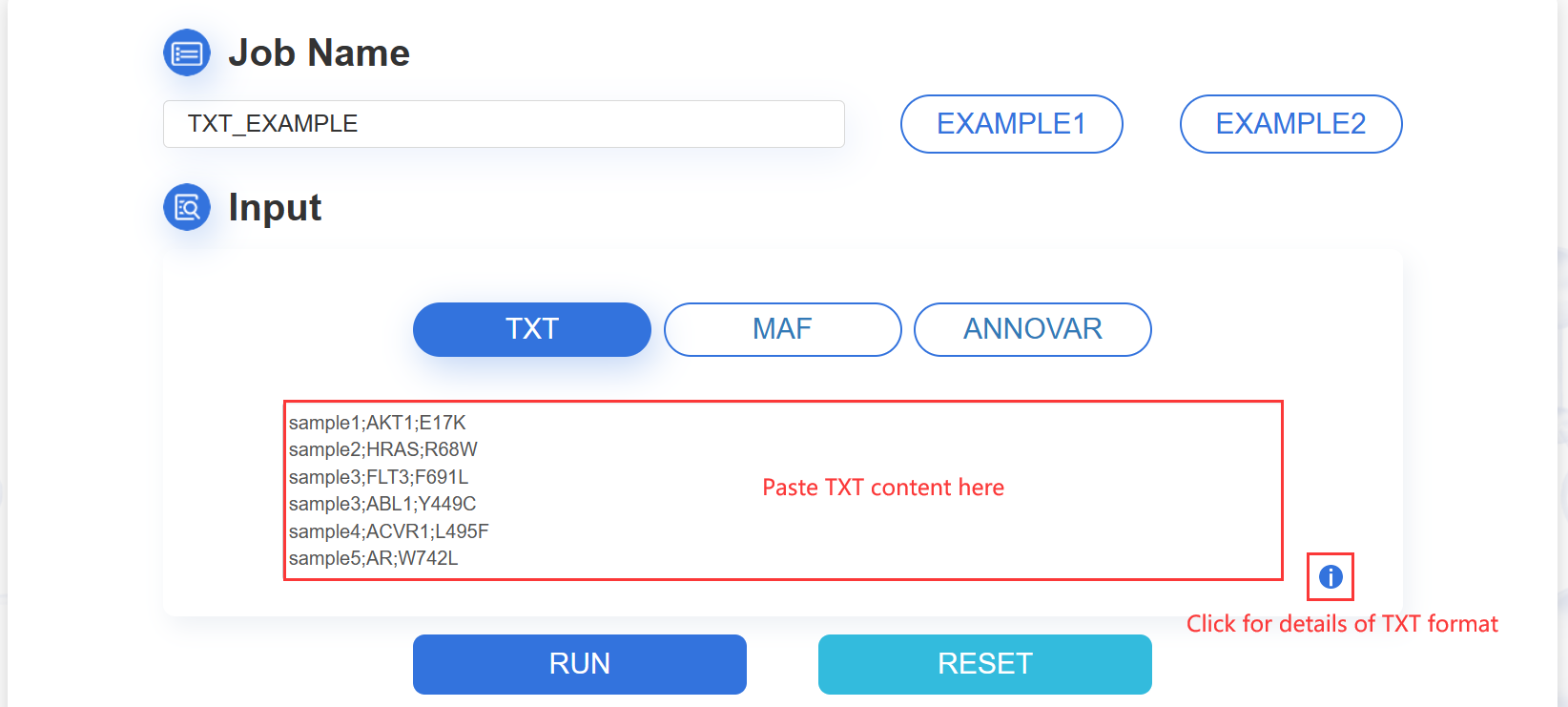
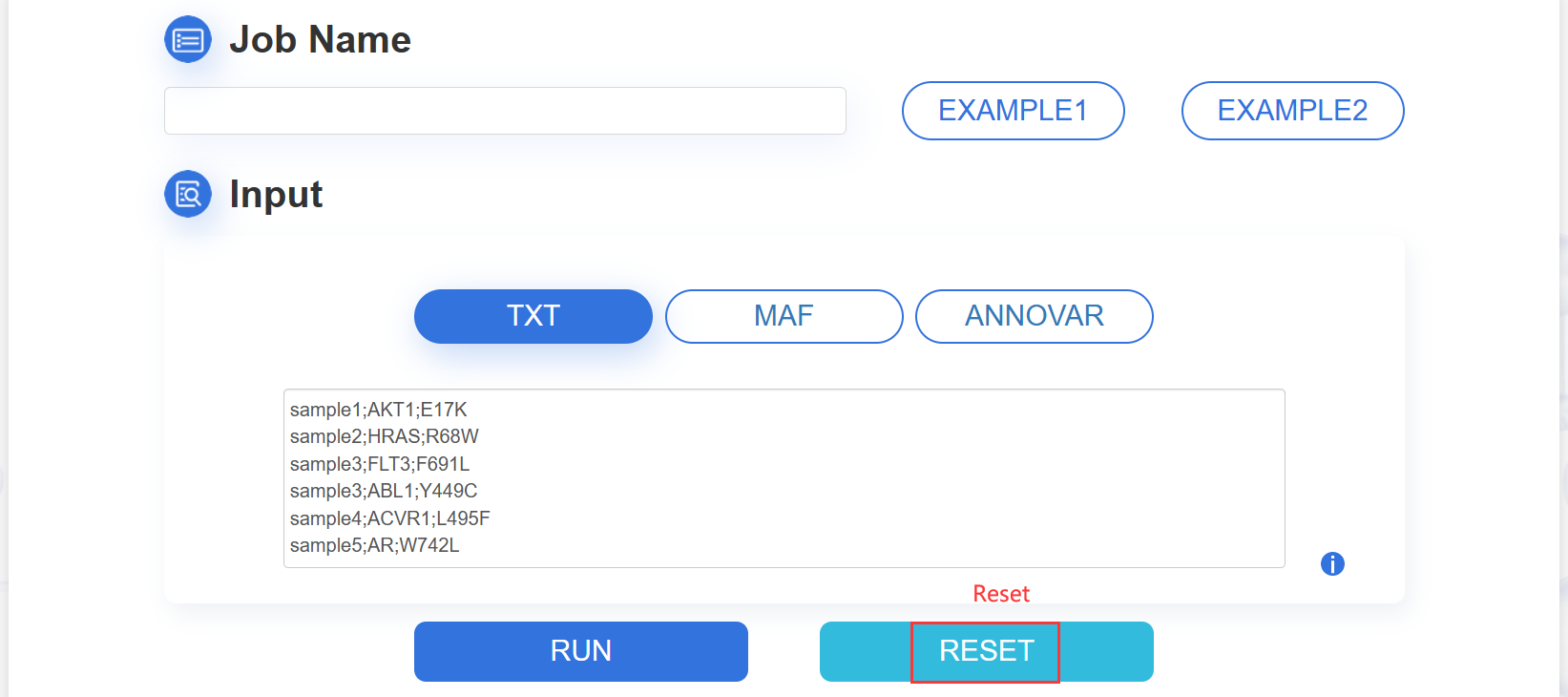
1.TXT:paste your mutation content into the text area:
The input mutation list contains three columns: Sample ID, Gene name and Mutation, which are separated by semicolon. The format are as following image:
ATTENTION: The input contents for prediction should be no more than 10M.
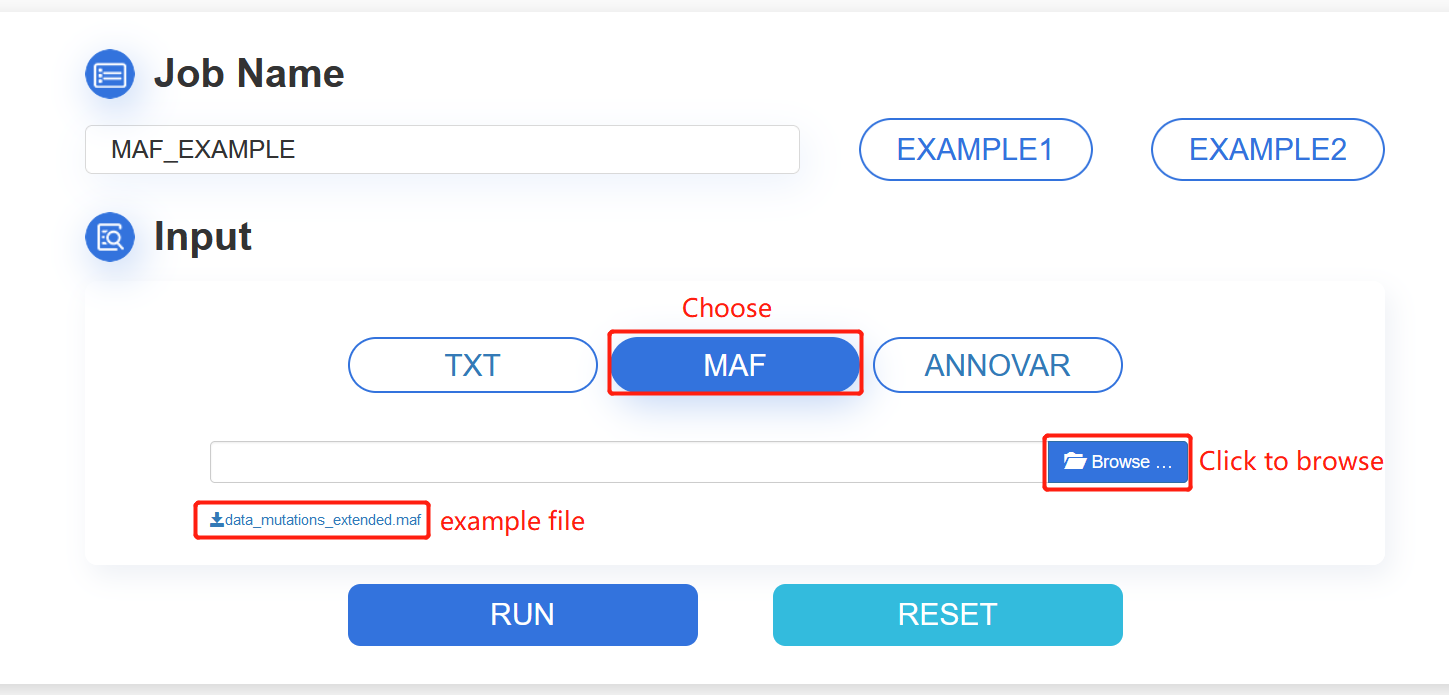
2. MAF: click the MAF button and browse your MAF file. Then, click the Browse button for file selecting. The file extension is allowed to be maf or txt. The file will be uploaded automatically.
A Mutation Annotation Format (MAF) file (.maf) is a human-readable and tab-delimited text file that lists mutations derived from sequencing cancer cohort samples. The format originates from The Cancer Genome Atlas (TCGA) project and has been widely accepted by researchers since the detected mutations in a MAF file can be easily viewed, edited or processed for custom mining. Each line in a MAF file represents a cancer mutation, which is annotated by many fields ranging from chromosome names to protein information. Column headers and ordering may sometimes vary between files of different sources.
In DeepAlloDriver, the MAF file is composed of four headers as follows: (1) "Hugo_Symbol" for gene symbol, e.g. BRAF; (2) "HGVSp_Short" for detected mutation, e.g. p.V600E; (3) "Variant_Classification" for translational effect of variant allele, e.g. Missense_Mutation; (4) "Tumor_Sample_Barcode" for cancer sample ID. More headers in MAF file are described in the GDC documentation of NIH (
https://docs.gdc.cancer.gov/Data/File_Formats/MAF_Format/ ).
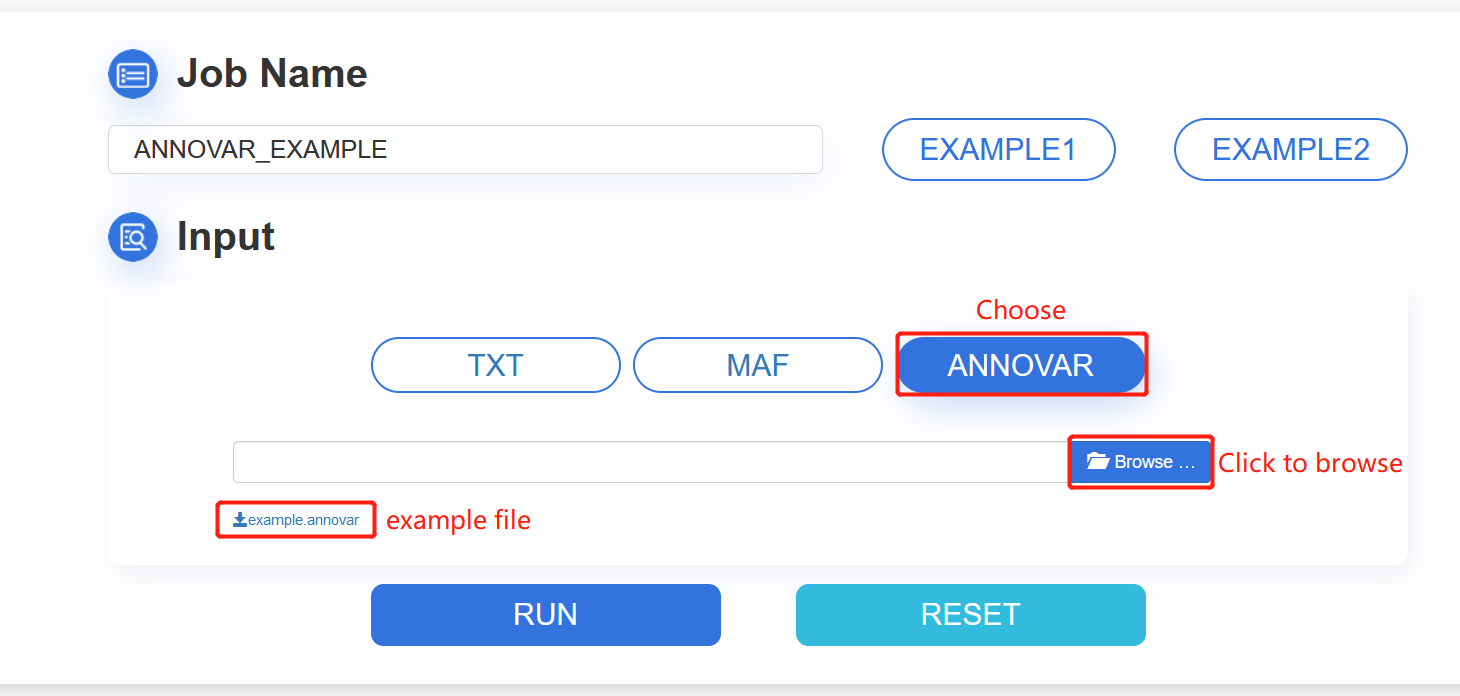
3. ANNOVAR: click the
ANNOVAR button and browse your ANNOVAR file. Then, click the
Browse button to choose a file. The file extension is allowed to be
annovar. The example annovar file can be downloaded
here. The file will be uploaded automatically.
ANNOVAR is a widely-used software to annotate genetic variants detected from diverse genomes. In DeepAlloDriver, a tab-separated annotation output file from ANNOVAR software are supported. The variant annotation file can be generated by the table_annovar.pl script in the program.
ATTENTION:The input files for prediction should should be no more than 10M.