Web Services
 |
ODORactor is an open access web server aimed at providing a platform for identifying odorant receptors (ORs) for small molecules and for browsing existing OR-ligand pairs. It enables the prediction of ORs from the molecular structures of arbitrary chemicals by integrating two individual functionalities: odorant verification and OR recognition. The prediction of the ORs for several odorants was experimentally validated in the study. In addition, ODORactor features a comprehensive repertoire of olfactory information that has been manually curated from literature. Therefore, ODORactor may provide an effective way to decipher olfactory coding and could be a useful server tool for both basic olfaction research in academia and for odorant discovery in industry. Enter |
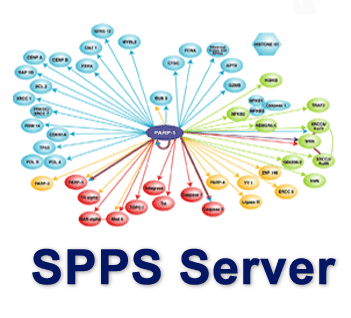 |
The molecular network sustained by different types of interactions among proteins is widely manifested as the fundamental driving force of cellular operations. Many biological functions are determined by the crosstalk between proteins rather than by the characteristics of their individual components. Thus, the searches for protein partners in global networks are imperative when attempting to address the principles of biology. We have developed a web-based tool "Sequence-based Protein Partners Search" (SPSS) to explore interacting partners of proteins, by searching over a large repertoire of proteins across many species. SPPS provides a database containing more than 40,000 protein sequences with annotations and a protein-partner search engine in two modes (Query1 and Query2). In addition, users can refine potential protein partner hits by using several filters and possible interactive network in the SPPS web server. SPPS provides a new type of tool to facilitate the identification of direct or indirect protein partners which may guide scientists on the investigation of new signaling pathways. Enter |
 |
The use of allosteric modulators as preferred therapeutic agents against classic orthosteric ligands has colossal advantages, including higher specificity, fewer side effects and lower toxicity. Therefore, the computational prediction of allosteric sites in proteins is receiving increased attention in the field of drug discovery. Allosite is a newly developed automatic tool for the prediction of allosteric sites in proteins of interest and is now available through a web server. Enter |










